What's this?
Accounts have been assigned only to users involved in the annotation process. If you believe you need an account, please contact Nicholas Heller at helle246@umn.edu.
What's this?
Accounts have been assigned only to users involved in the annotation process. If you believe you need an account, please contact Nicholas Heller at helle246@umn.edu.
The 2023 Kidney and Kidney Tumor Segmentation challenge (abbreviated KiTS23) is a competition in which teams compete to develop the best system for automatic semantic segmentation of kidneys, renal tumors, and renal cysts. It is the third iteration of the KiTS challenge after having taken place in 2019 and 2021.
Kidney cancer is diagnosed in more than 430,000 individuals each year, accounting for roughly 180,000 deaths [1]. Kidney tumors are found in an even greater number each year, and in most circumstances, it's not currently possible to radiographically determine whether a given tumor is malignant or benign [2]. Even among tumors presumed to be malignant, many appear to be slow-growing and indolent in nature, which has given rise to "active surveillance" as an increasingly popular management strategy for small renal masses [3]. That said, the progression to metastatic disease is a very serious concern, and there is a significant unmet need for systems that can objectively and reliably characterize kidney tumor images for stratifying risk and for pedicting treatment outcomes.
For nearly half a decade, KiTS has maintained and expanded upon a publicly-available, multi-institutional cohort of hundreds of segmented CT scans showing kidney tumors, along with comprehensive anonymized clinical information about each case [4]. This dataset has not only served as a high-quality benchmark for 3D semantic segmentation methods [5,6], but also as a common resource for translational research in kidney tumor radiomics [7,8].
For the third time, KiTS is inviting the greater research community to participate in a competition to develop the best automatic semantic segmentation system for kidney tumors. This year's competition features an expanded training set (489 cases), a fresh never-before-used test set (110 cases), and the addition of cases in the nephrogenic contrast phase, whereas previously all cases were in late arterial. We are excited to see how modern approaches can perform in this more diverse and challenging -- and also more broadly-applicable context.
| April 14 | Training Dataset Release |
| July 14 | Deadline for Short Paper |
| July 21 - 28 | Submissions Accepted |
| July 31 | Results Announced |
| October 8 | Satellite Event at MICCAI 2023 |
The KiTS23 cohort includes patients who underwent cryoablation, partial nephrectomy, or radical nephrectomy for suspected renal malignancy between 2010 and 2022 at an M Health Fairview medical center. A retrospective review of these cases was conducted to identify all patients who had undergone a contrast-enhanced preoperative CT scan that includes the entirety of all kidneys.
Each case's most recent contrast-enhanced preoperative scan (in either corticomedullary or nephrogenic phase) was segmented for each instance of the following semantic classes.
The dataset is composed of 599 cases with 489 allocated to the training set and 110 in the test set. Many of these in the training set were used in previous challenges:
The 110 cases in the KiTS23 test set, on the other hand, have not yet been used for any challenge.

Fig. 3: A visualization showing the largest axial slice of each tumor in the dataset overlayed onto one image
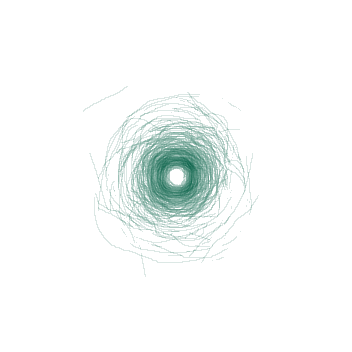
Fig. 4: A visualization showing the boundary between the tumor and the kidney on the the tumor's largest axial slice overlayed onto one image
Just like KiTS21, we have decided to perform our image annotations on a web-based platform in full view of the public. We hope that this will encourage participants to scrutinize this process and dataset in greater detail, and perhaps reproduce something similar for their own work.
The process for KiTS23 is very similar to what was described for KiTS21. The primary difference is that we have decided to skip the "guidence" step, and instead have the team of trainees directly perform the "delination" step, and in so doing, each region of interest is segmented only once. We made this decision based on the KiTS21 finding that the three delineations per ROI offered only marginal performance boosts to methods that made use of it, if any at all, and therefore does not seem to represent a wise allocation of scarce annotation resources.
If you choose to browse the annotation (and we hope you do!), you will see a card for each case that indicates its status in the annotation process. An example of this is shown in Fig. 5. The meaning of each symbol is as follows:
When you click on an icon, you will be taken to an instance of the ULabel Annotation Tool where you can see the raw annotations made by our annotation team. Only logged-in members of the annotation team can submit their changes to the server, but you may make edits and save them locally if you like. The annotation team's progress is synced with the KiTS23 GitHub repository about once per week.
It's important to note the distinction between what we call "annotations" and what we call "segmentations". We use "annotations" to refer to the raw vectorized interactions that the user generates during an annotation session. A "segmentation," on the other hand, refers to the rasterized output of a postprocessing script that uses "annotations" to define regions of interest.
We placed members of our annotation team into three categories:
Broadly, our annotation process is as follows.
Our postprocessing script uses thresholds and fairly simple heuristic-based geometric algorithms. Its source code is available on the KiTS23 GitHub repository under /kits23/annotation/postprocessing.py.
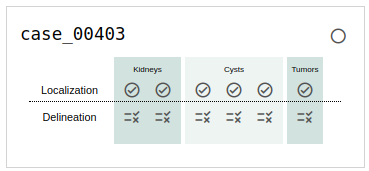
Fig. 5: An example of a case in progress as it's listed on the browse page.
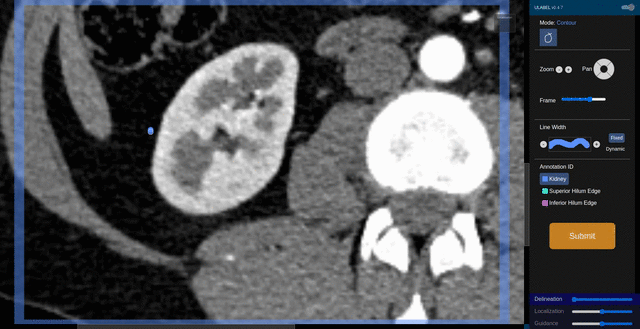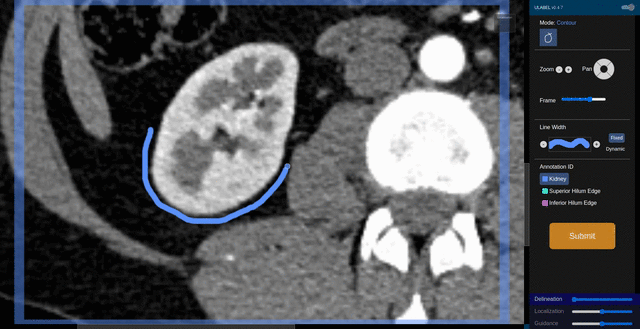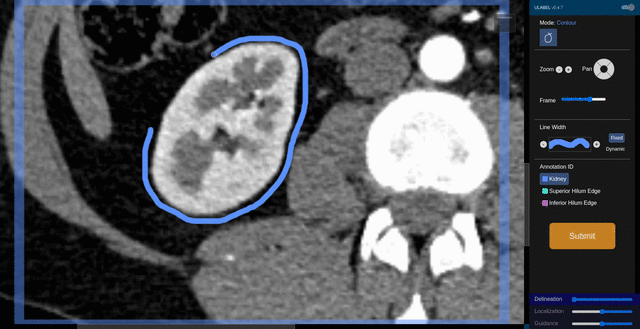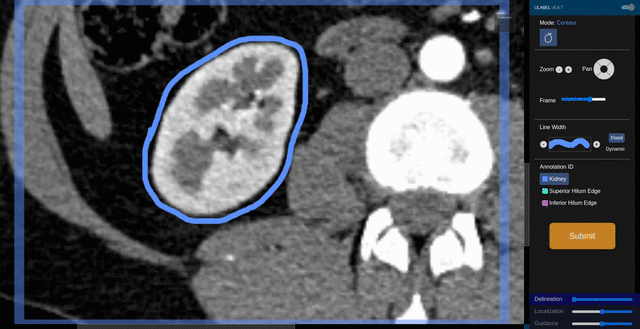
Fig. 6: An animation showing a kidney delineation of one axial slice. You'll notice that the contour does not keep perfectly snug to the ROI. This is because the postprocessing can easily remove the included perinephric fat using a Hounsfield Unit threshold.
Like KiTS21 and KiTS19 before it, we will be using "Hierarchical Evaluation Classes" (HECs) rather than each ROI alone. The HECs are as follows:
For a more detailed discussion of why we use HECs, please see the explanation from KiTS21.
We will also be using the same two metrics that we used in KiTS21:
These will be computed for every HEC of every case of the test set and the metrics will be averaged over each HEC. The teams will then be ranked based on both average metrics. The final leaderboard place will be determined by averaging the leaderboard places between the two rankings, sometimes called "rank-then-aggregate". In the case of any ties, average Sørensen-Dice value on the "Tumor" HEC will be used as a tiebreaker. An implementation of these metrics and ranking procedure can be found at /kits23/evaluation/.
The winning team will be awarded $5,000 USD after their containerized solution has been verified
This project is a collaboration between the University of Minnesota Robotics Institute (MnRI), the Helmholtz Imaging at the German Cancer Research Center (DKFZ), and the Cleveland Clinic's Urologic Cancer Program.
 This challenge was made possible by scholarships awarded by Climb 4 Kidney Cancer (C4KC), an organization dedicated to advocacy for kidney cancer patients and the advancement of kidney cancer research.
This challenge was made possible by scholarships awarded by Climb 4 Kidney Cancer (C4KC), an organization dedicated to advocacy for kidney cancer patients and the advancement of kidney cancer research.
 This work was also supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA225435.
This work was also supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA225435.
